Modeling of Oxygen Metabolism in 3D Cell Constructs with LiveLink™ for MATLAB®
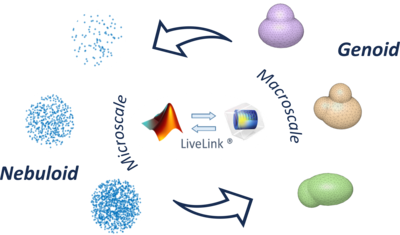
In silico models can be exploited to predict cellular dynamics, such as resource uptake, migration and proliferation. Continuum modelling strategies such as finite element (FE) simulations are amongst the most common, but cannot exhaustively describe the metabolic consumption of cells, since encapsulated cells in 3D structures are composed of discrete consuming units within non consuming extracellular space. Alternatively, one can implement an agent-based approach. Here, cells are considered as autonomous domains described by predefined state variables. Such models allow evaluating macroscopic changes of cellular constructs (e.g., reshaping, expansion) and are mostly based on cell-cell or cell-environment mechanical interactions. Exploiting LiveLink™ for MATLAB®, here we present a multi-scale approach, from the micro -(cells) up to the macro- (the whole construct) scale, to empower FE models as tools for evaluating metabolic dynamics in three-dimensional (3D) cell constructs (e.g., organoids), for their application in the design of physiologically relevant cellular systems. In particular, our method relies on exploiting a so-called Nebuloid (i.e., an agent-based model to account for cell scale metabolic dynamics) to describe macroscale aspects (e.g., mass transport, morphological changes).
Unlike the assumption of homogeneous cell density usually reported for continuum-like models, the Nebuloid is modelled as a point cloud simulating the heterogeneous spatial distribution observed in hepatocyte-laden spheroids. It also allows cell-specific tuning of metabolic parameters based on our experimental measurements of oxygen consumption kinetics according to the Michaelis-Menten formulation. Cellular motility and proliferation are implemented according to Boolean constraints on the concentration fields in the surrounding continuum, which can in turn be reshaped depending on cell dynamics. The Transport of Diluted Species interface is used for solving the diffusion-reaction equation at the steady state. The outcomes from our model were compared to those obtained using traditional continuum domains, for corresponding geometries. The Nebuloid method computes significantly lower construct metabolic rates, in line with experimental observables. We are currently integrating our Nebuloid model with the LiveLink™ for MATLAB® exploiting genetic algorithms (GAs) to predict the macroscale morphology of 3D cell constructs. Briefly, the oxygen consumption and surface energy of a random population of constructs laden with cells with different morphologies is generated. Each of them is simulated using the Nebuloid model and scored through a fitness function (FF), defined ad hoc to account for the biophysical constraints of interest. Then, individuals yielding optimal morphologies are identified by minimizing the FF, and a new generation is obtained randomly combining their geometrical features. The process is iterated until the FF value stabilizes within a threshold of tolerance over consecutive generations or the predefined maximum number of generations is reached.
To conclude, LiveLink™ for MATLAB® is used to model micro, meso- and macroscale dynamics occurring in cellular systems with FE analysis, allowing more reliable predictions of oxygen metabolism by cells as well as determination of optimal shapes of cell-laden constructs. Future developments will include experimental validation of the approach.
Download
- Mancini_6271_poster.pdf - 0.79MB
